ConfID
ConfID
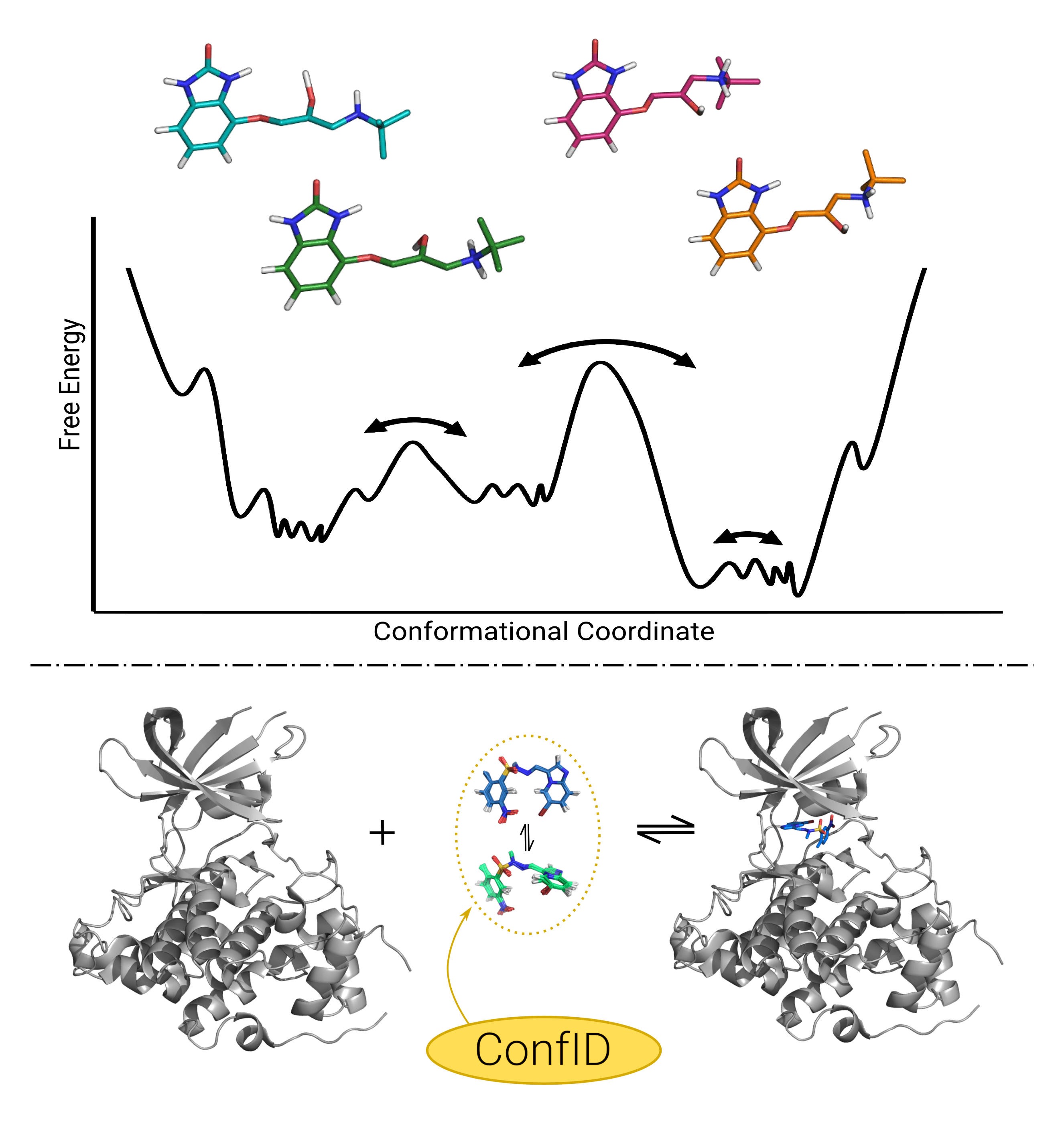
Conformational Identifier for drug-like moleculesConformational generation is a recurrent challenge in early phases of drug design, mostly due to the task of making sense between the number of conformers generated and their relevance for biological purposes.
In this sense, ConfID, a Python-based computational tool, was designed to identify and characterize conformational populations of drug-like molecules sampled through molecular dynamics simulations.
By using molecular dynamics (MD) simulations (and assuming accurate parameters are used), ConfID can identify all conformational populations sampled in the presence of solvent and quantify their relative abundance, while harnessing the benefits of MD and calculating time-dependent properties of each conformational population identified.
To contact us, please drop an email to bigrisci@inf.ufrgs.br and mdpoleto@vt.edu.
What is ConfID?
It is a Python-based computational tool designed to identify and characterize conformational populations of small molecules sampled through molecular dynamics simulations.
ConfID was developed by:
Bruno I. Grisci - PhD student (Institute of Informatics - UFRGS)
Marcelo D. Polêto - Postdoctoral Associate (Biochemistry Department - VirginiaTech)
Marcio Dorn - Associate Professor (Institute of Informatics - UFRGS)
Hugo Verli - Associate Professor (Center of Biotechnology - UFRGS)
To which problem ConfID was designed for?
Genetic algorithms and knowledge-based approaches have been employed to study molecular flexibility. However, these methods are usually based on crystallographic information, and their calculations are made in vacuum or with implicit solvent and do not take into account the influence of explicit solvent molecules on conformational preferences.
By using structural information gathered from MD simulations (and assuming accurate parameters are used), ConfID can identify all conformational populations of drug-like molecules sampled in the presence of solvent and quantify their relative abundance, while harnessing the benefits of MD and calculating time-dependent properties of each conformational population.
Tutorials
Frequently Asked Questions
Do you have any questions? Take a look at our FAQ.
Publications
There are some papers already using ConfID! These are some:
- Molecular dynamic simulation: Conformational properties of single-stranded curdlan in aqueous solution FENG, X.; LI, F.; DING, M.; ZHANG, R.; SHI, T. Carbohydrate Polymers, v. 250, p. 116906, 2020.
- The Lazy Life of Lipid-Linked Oligosaccharides in All Life Domains ARANTES, P. R.; PEDEBOS, C.; POLÊTO, M. D.; POL-FACHIN, L.; VERLI, H. Journal of Chemical Information and Modeling, v. 60, p. 631–643, 2020.
- An Unusual Intramolecular Halogen Bond Guides Conformational Selection TESCH, R.; BECKER, C.; MÜLLER, M. P.; BECK, M. E.; QUAMBUSCH, L.; GETLIK, M.; LATEGAHN, J.; UHLENBROCK, N.; COSTA, F. N.; POLÊTO, M. D.; PINHEIRO, P. S. M.; RODRIGUES, D. A.; SANT'ANNA, C. M. R.; FERREIRA, F. F.; VERLI, H.; FRAGA, C. A. M.; RAUH, D. Angewandte Chemie International Edition, v. 57, p. 9970-9975, 2018.
- Development of GROMOS-Compatible Parameter Set for Simulations of Chalcones and Flavonoids ARANTES, P. R.; POLÊTO, M. D.; JOHN, E. B. O.; PEDEBOS, C.; GRISCI, B. I.; DORN, M.; VERLI, H. The Journal of Physical Chemistry B, v. 123, p. 994-1008, 2019.
Citing ConfID
If you use ConfID in a scientific publication, we would appreciate citations to the following paper:
BibTeX
@article{10.1093/bioinformatics/btaa130,
author = {Polêto, M D and Grisci, B I and Dorn, M and Verli, H},
title = "ConfID: an analytical method for conformational characterization of small molecules using molecular dynamics trajectories",
journal = {Bioinformatics},
year = {2020},
month = {02},
issn = {1367-4803},
doi = {10.1093/bioinformatics/btaa130},
url = {https://doi.org/10.1093/bioinformatics/btaa130},
note = {btaa130},
eprint = {https://academic.oup.com/bioinformatics/advance-article-pdf/doi/10.1093/bioinformatics/btaa130/32677172/btaa130.pdf},
}