BARRA:CuRDa
BARRA:CuRDa
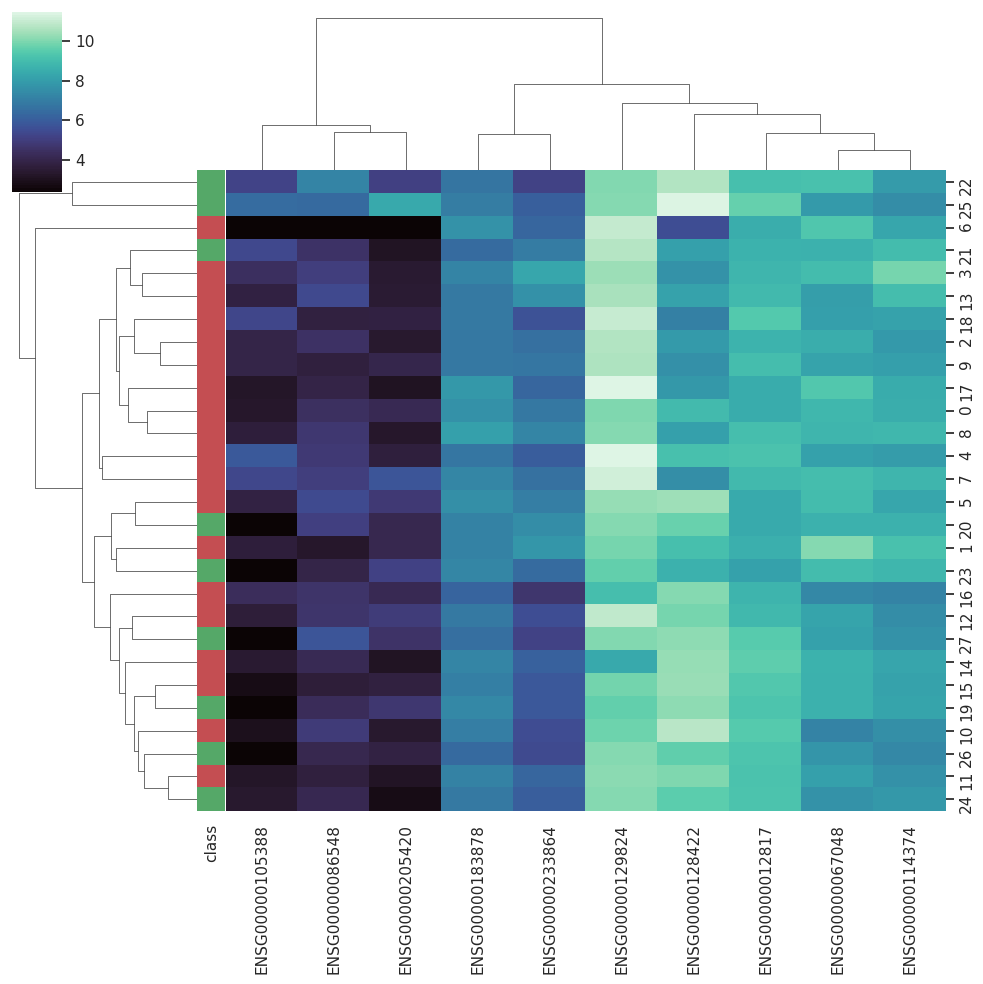
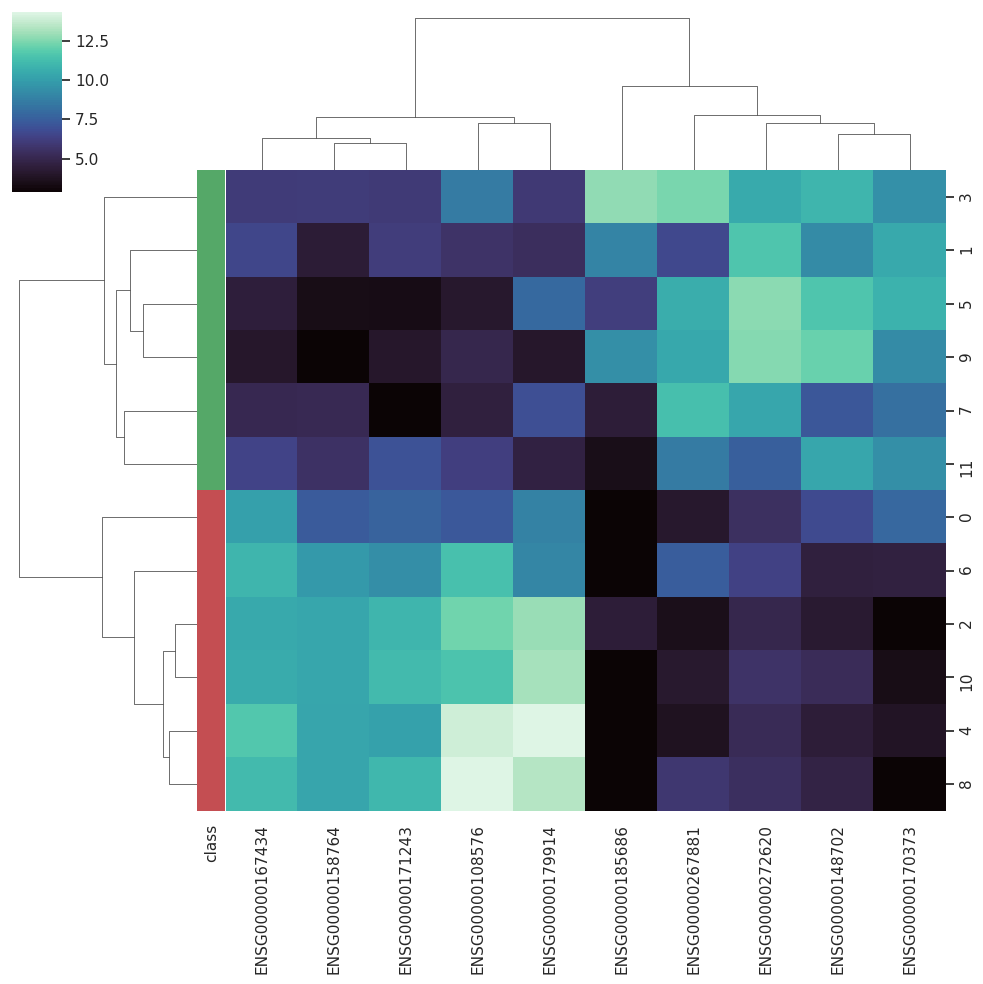
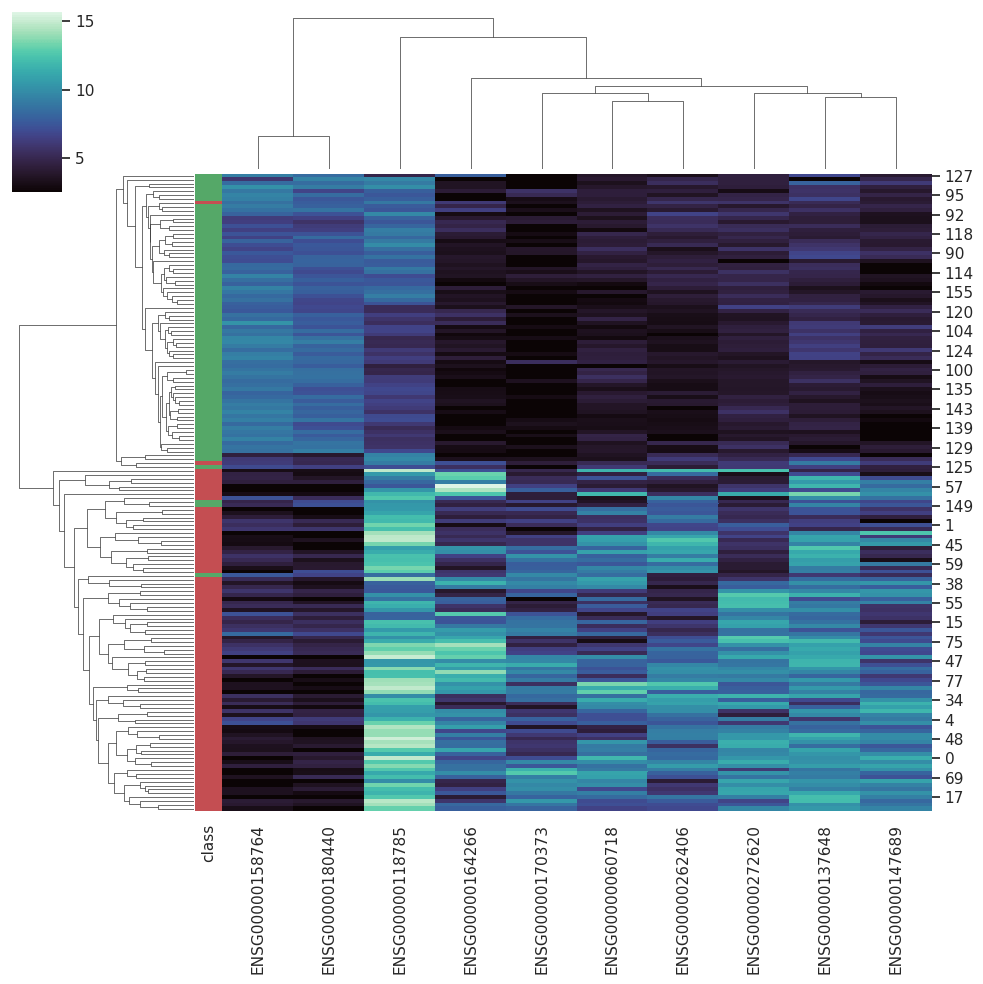
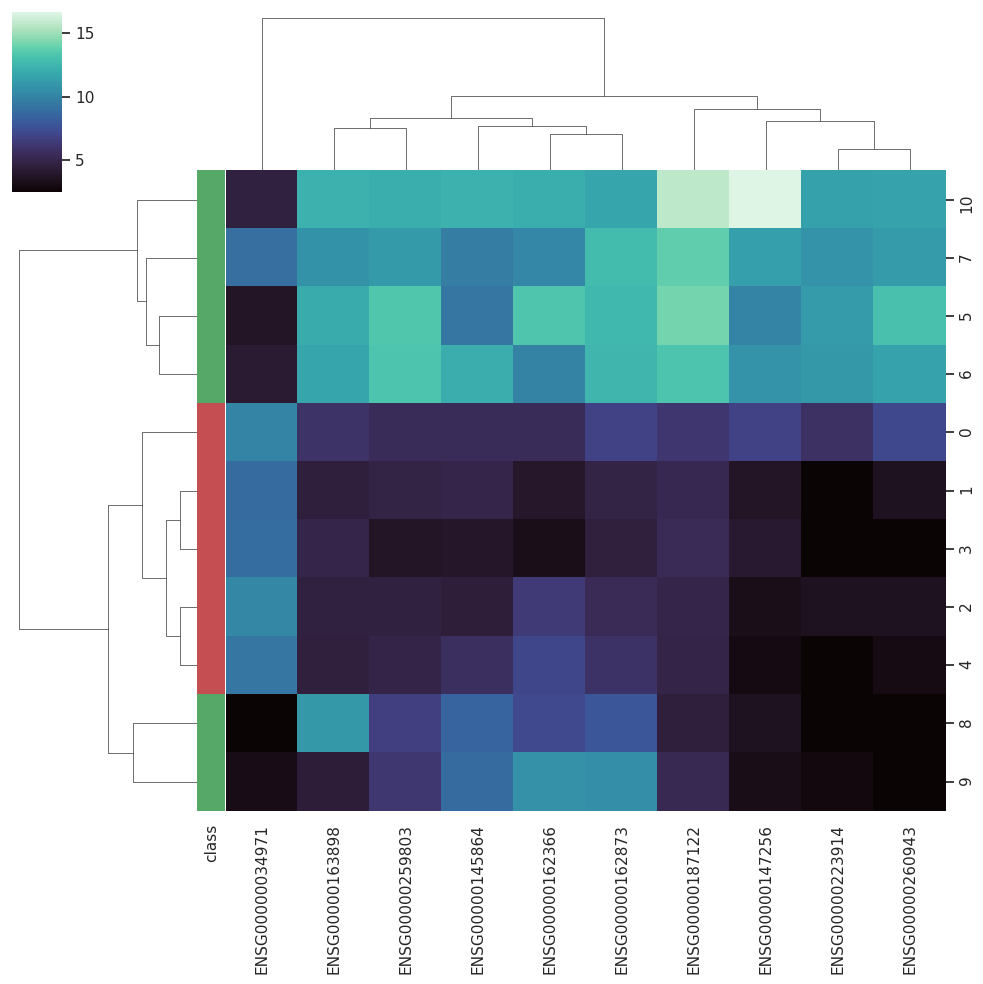
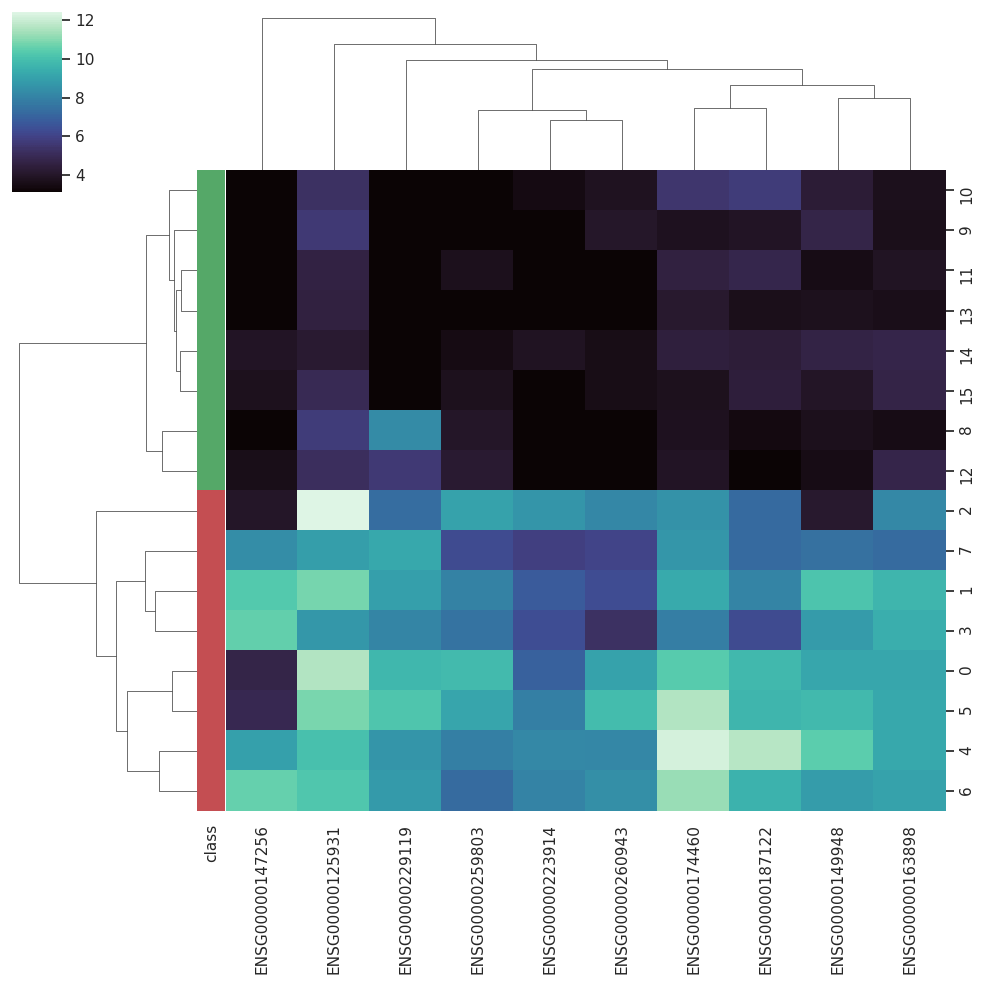
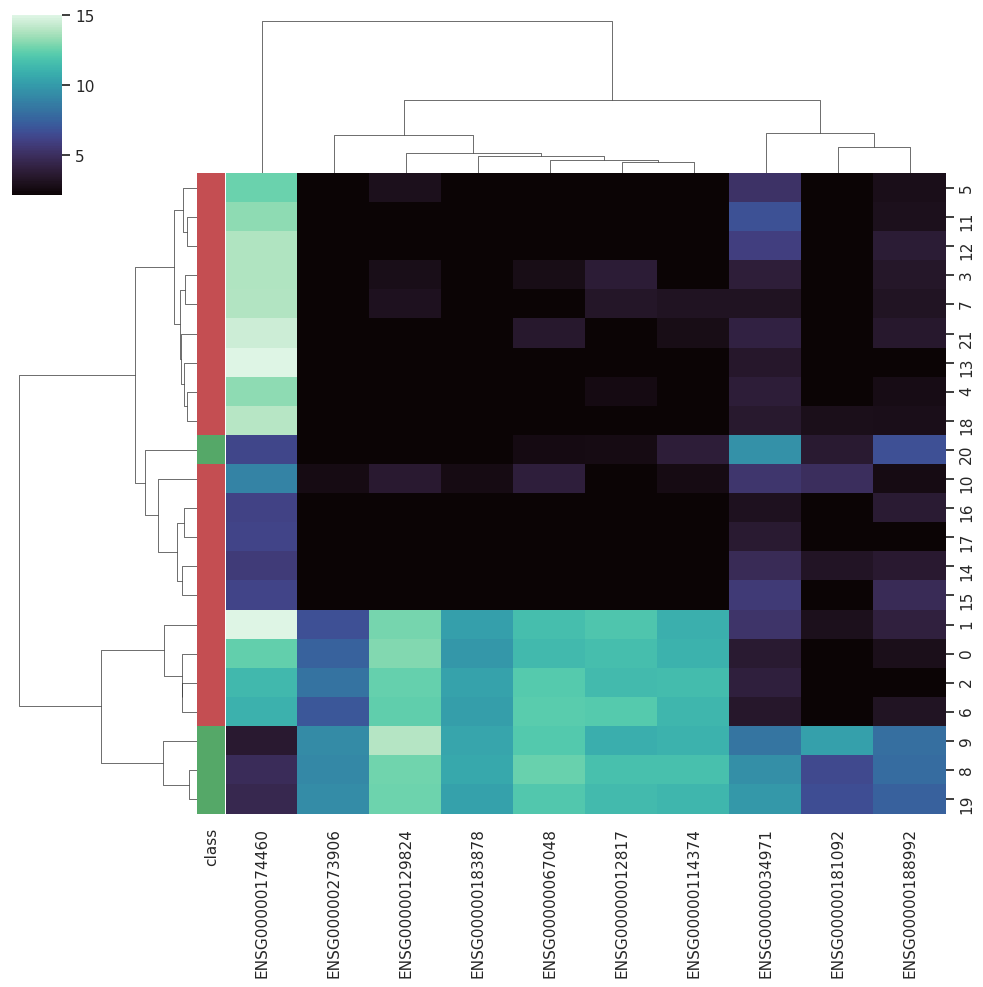
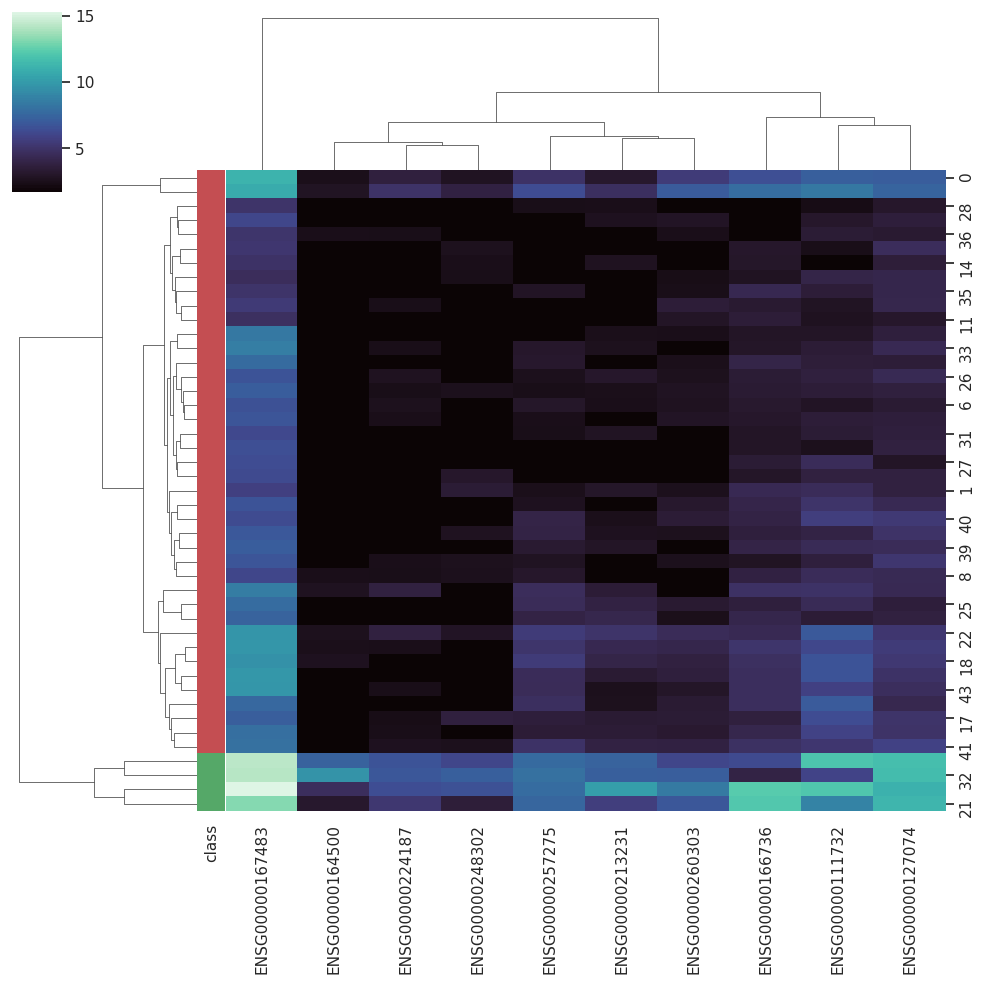
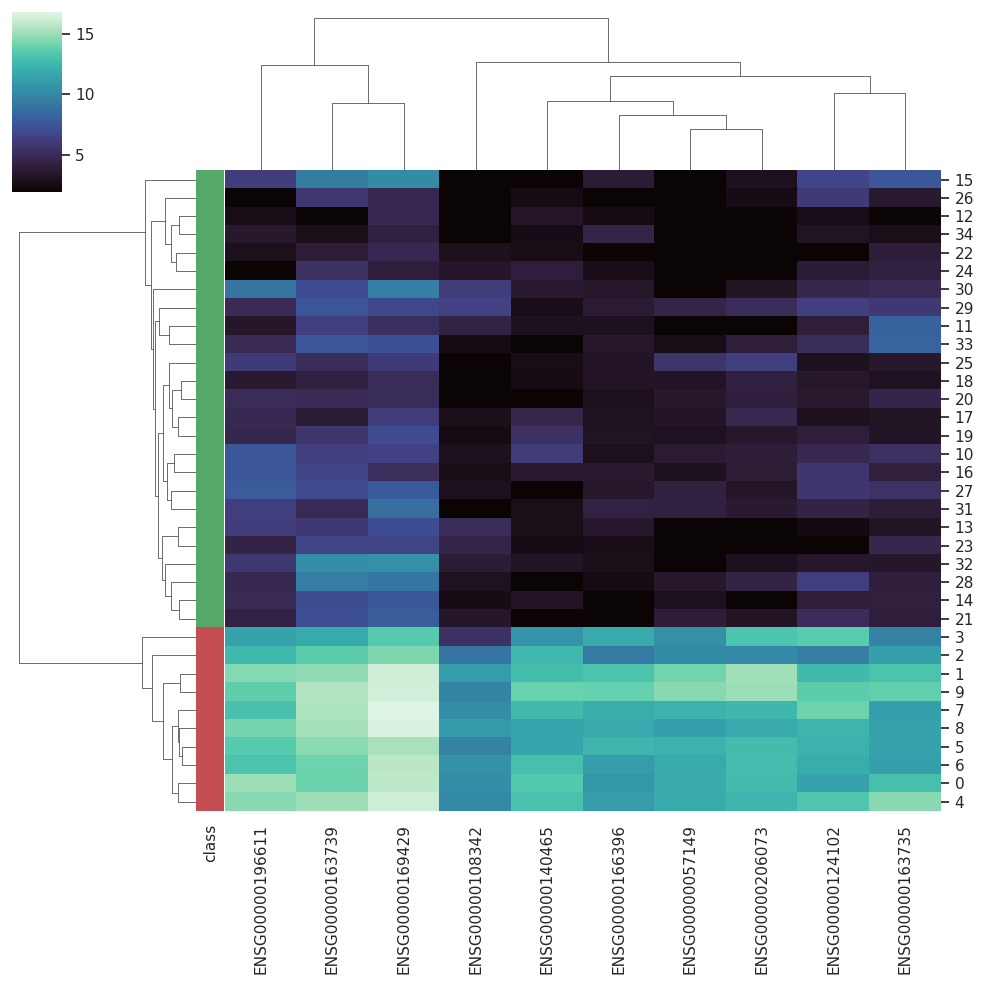
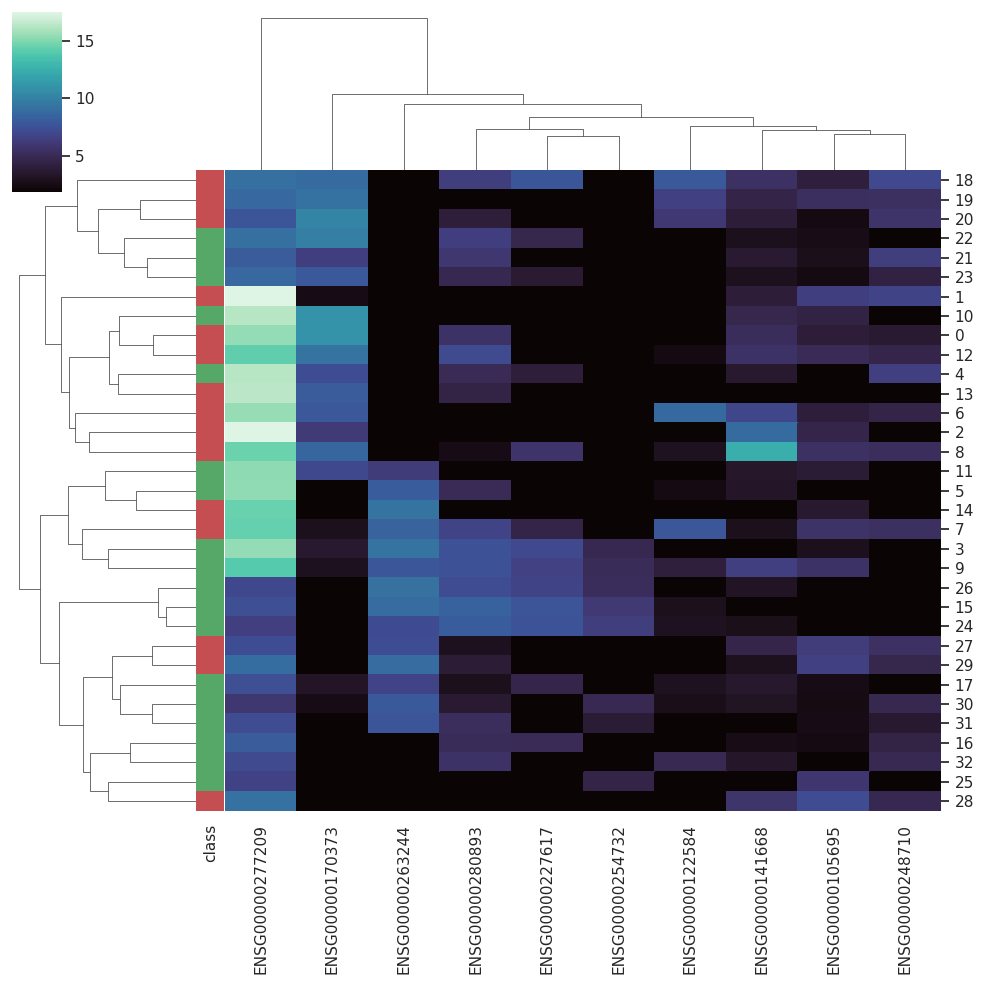
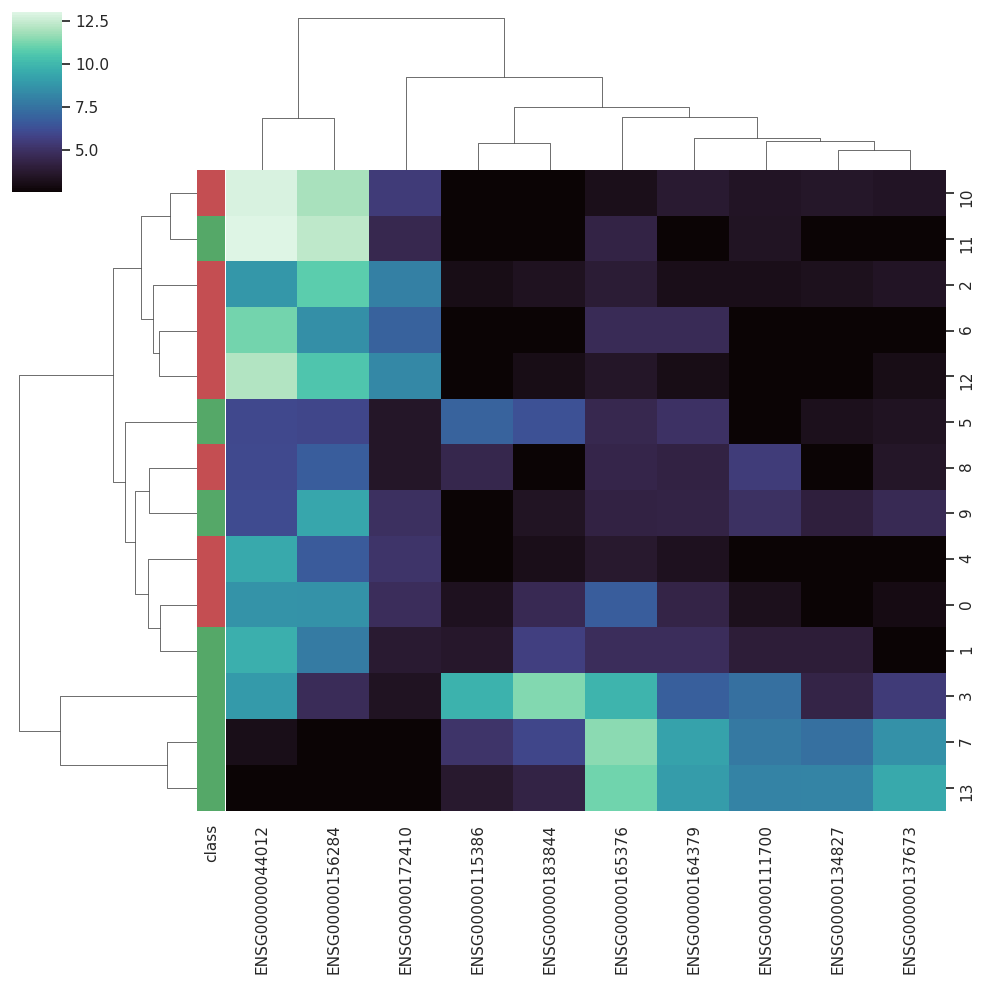
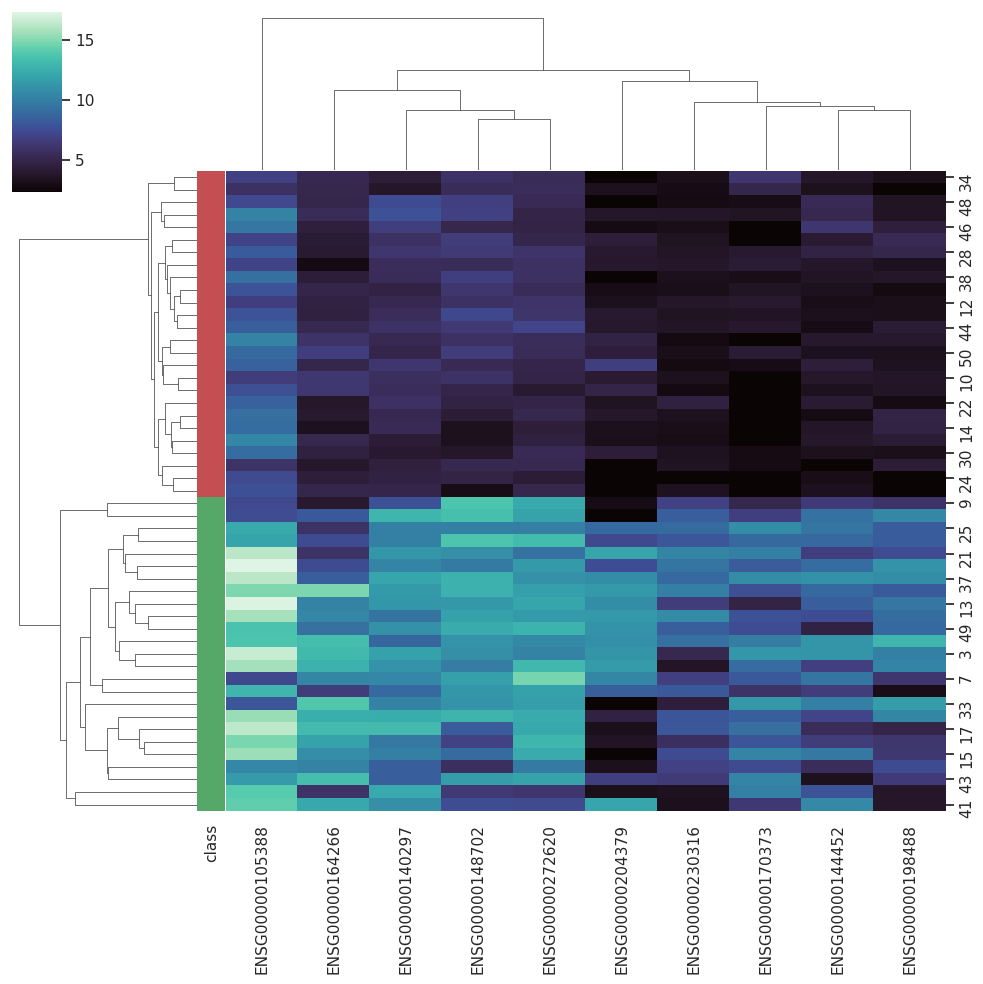
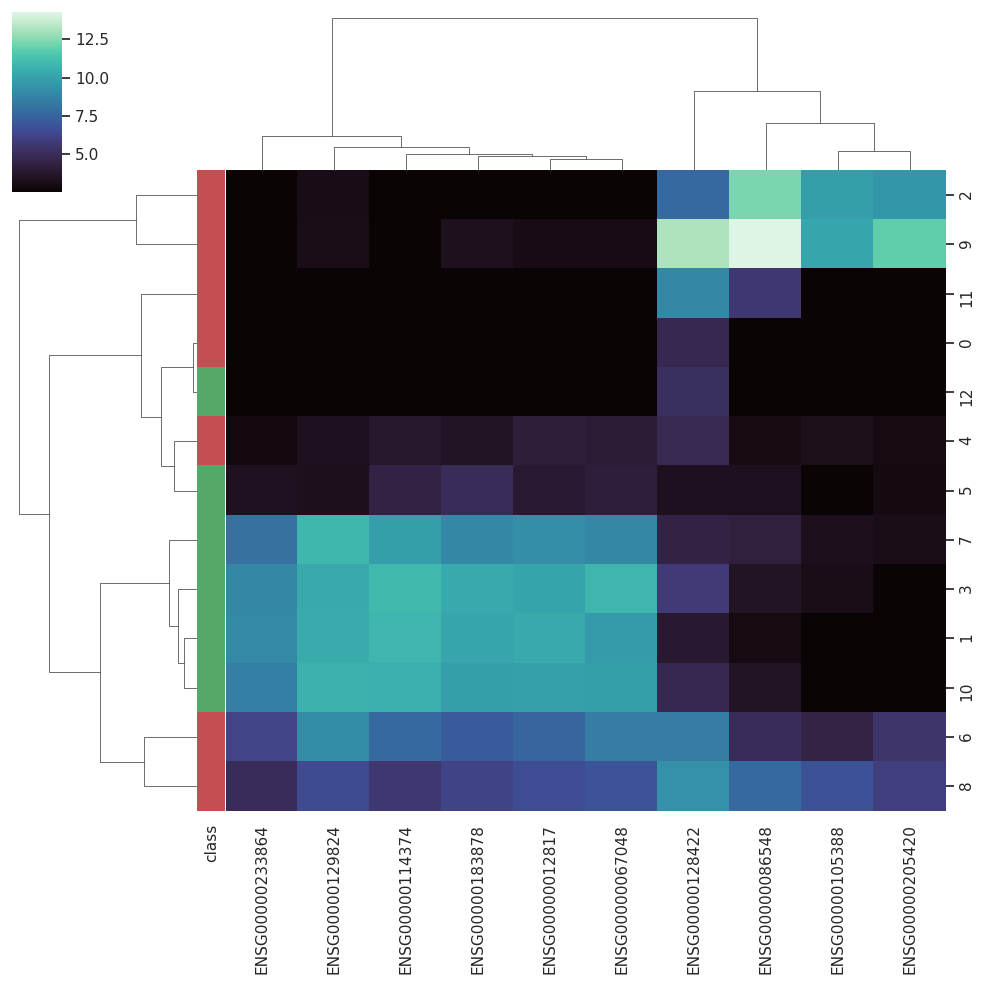
A Curated RNA-seq Database for Cancer ResearchRNA-seq is gradually becoming the dominating technique employed to access the global gene expression in biological samples, allowing more flexible protocols and robust analysis. However, the nature of RNA-seq results im-pose new data-handling challenges when it comes to computational analysis. With the increasing employment of machine learning techniques in biomedical sciences, databases that could provide curated datasets treated with state-of-the-art approaches already adapted to machine learning protocols become essential for testing new algorithms.
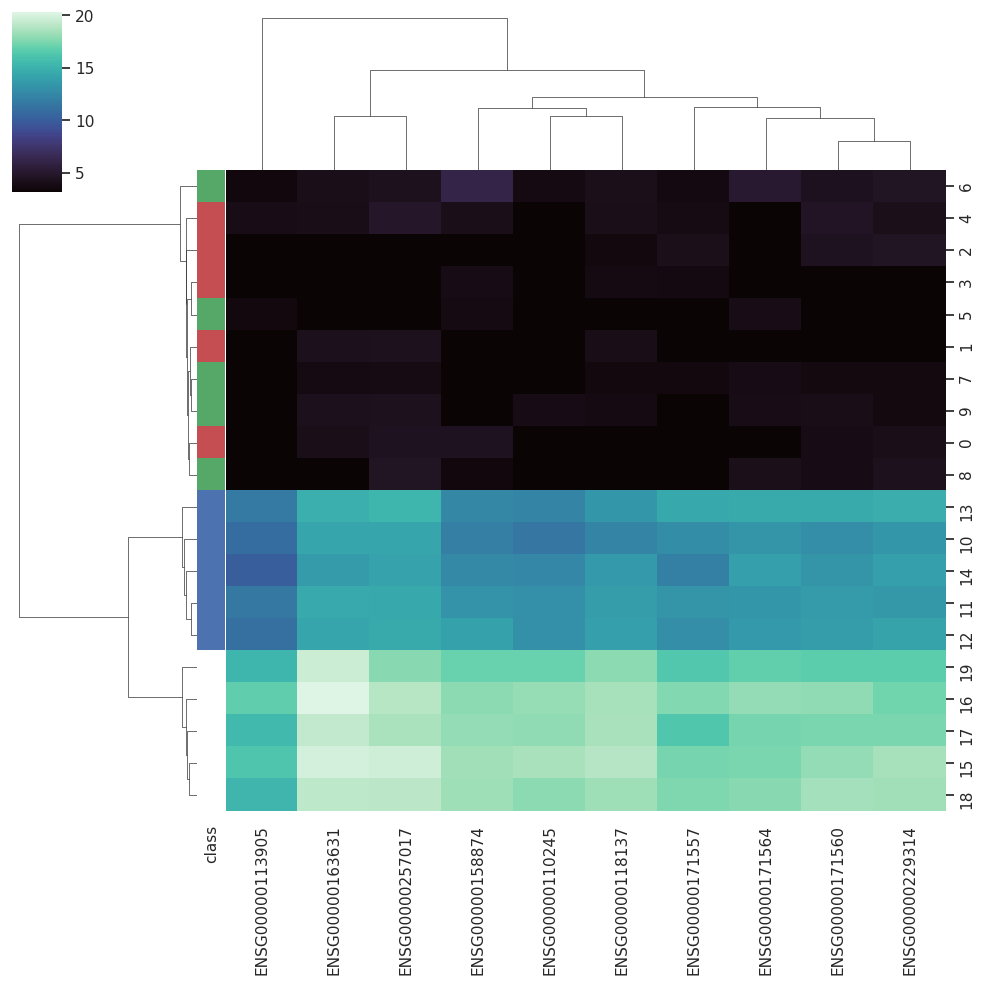
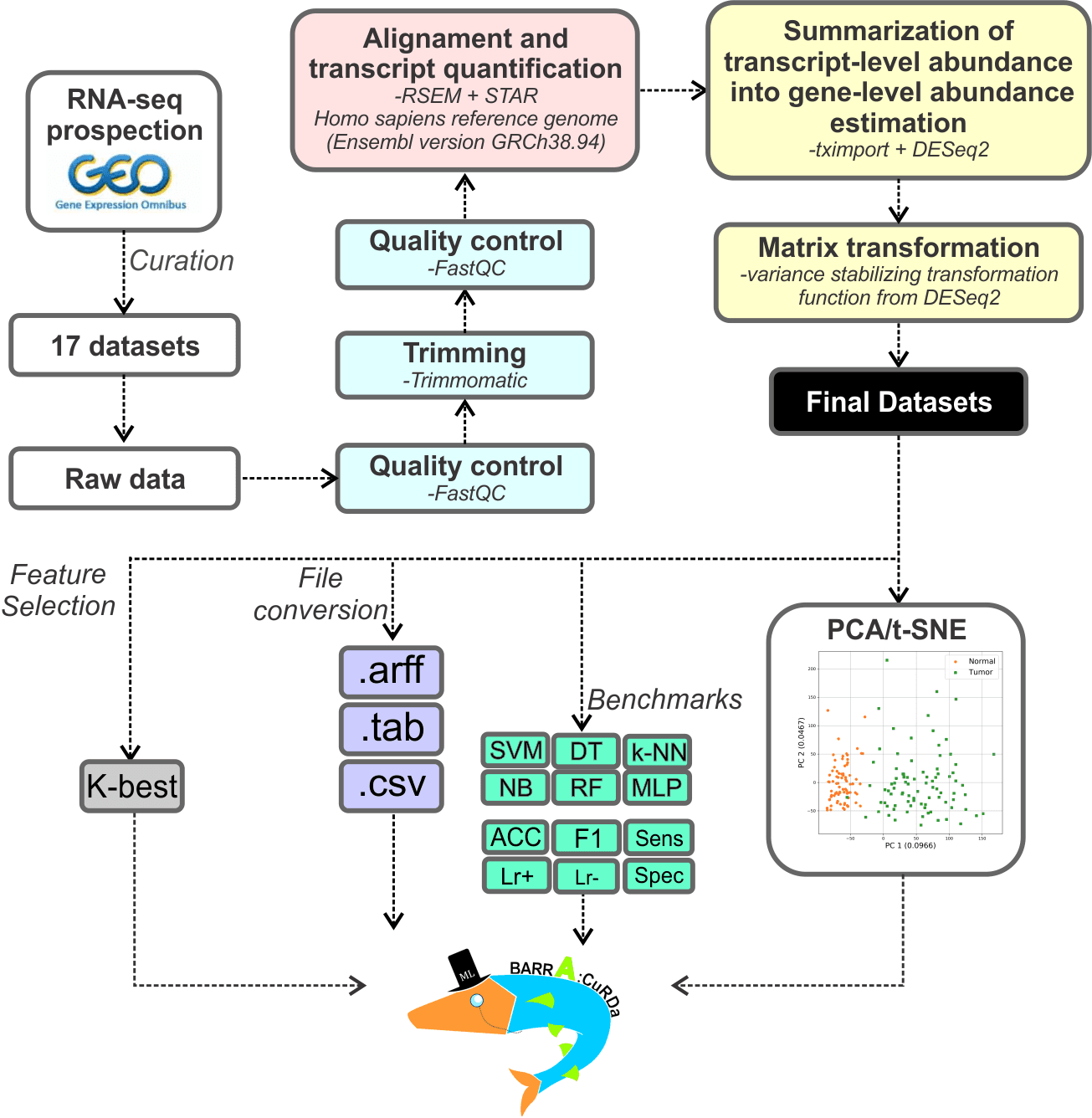
BARRA:CuRDa is composed of 17 handpicked RNA-seq datasets for Homo sapiens gathered from the Gene Expression Omnibus (GEO), using rigorous filtering criteria. All datasets were individually submitted to sample quality analysis, removal of low-quality bases, artifacts from the experimental process, removal of ribosomal RNA, and transcript level abundance. Moreover, like its sister database, all datasets were tested using analyses destined to provide a base knowledge of each dataset's characteristics, with the addition of new metrics.
How to CIte
If you use BARRA:CuRDa in a scientific publication, we would appreciate citations to the following paper:
BibTeX
@article{feltes:2021,
author = {Feltes, Bruno Cesar and Poloni, Joice De Faria and Dorn, Marcio},
doi = {10.1089/cmb.2020.0463},
journal = {Journal of Computational Biology},
number = {9},
pages = {931--944},
title = {{Benchmarking and Testing Machine Learning Approaches with BARRA:CuRDa, a Curated RNA-Seq Database for Cancer Research}},
url = {https://doi.org/10.1089/cmb.2020.0463},
volume = {28},
year = {2021}
}
Workflow
Datasets