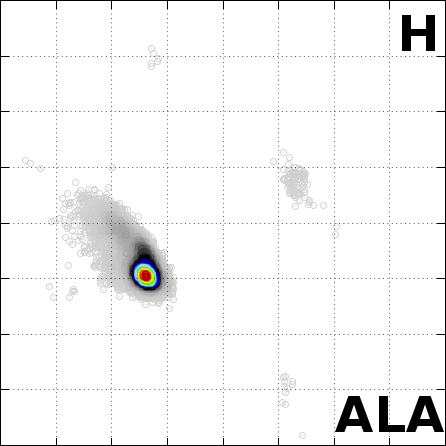
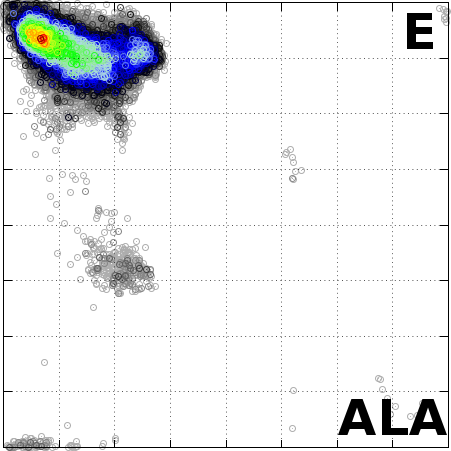
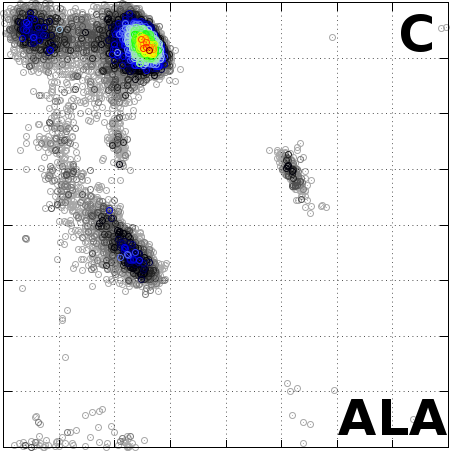
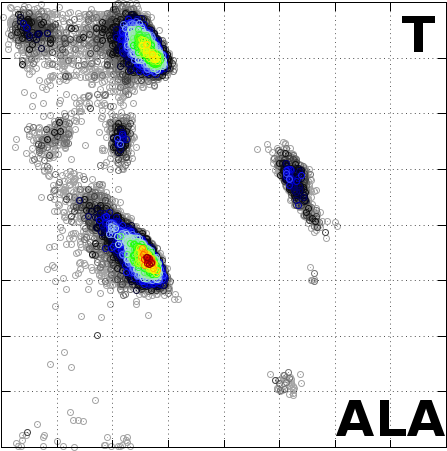
The Angle Probability List (APL) represents the normalized
frequency of observed pairs of amino acid residues and
secondary structure in the Protein Data Bank. We combine
the conformational preferences of amino acid residues
(AA, torsion angles) in proteins with their secondary
structure information (SS).
We selected a set of 6,650 protein structures
from PDB. All 3-D protein structures were experimentally
determined by X-ray diffraction with resolution ≤ 2.0Å
and stored in PDB until December 2014. We remove all
structures with R-factor greater than 0.2. If homologous
protein chains with sequence identity at most 30% were
found, only one of them was retained. We select only
amino acid residues with b-factor ≤ 30Å2 and occupancy
equal to 1. Similar parameters to filter PDB data were
used before by Hovmoller and Ohlson (2002).
For more details please contact us:
mdorn@inf.ufrgs.br
Download
APL Files (15.4 MB)Please cite APL as shown below:
DOI BibTexBORGUESAN, B.; BARBACHAN e SILVA, M.; GRISCI, B. I.; INSTROZA-PONTA, M.; DORN, M. APL: an Angle Probability List to improve knowledge-based metaheuristics for the three-dimensional protein structure prediction. Computational Biology and Chemistry (Print), v. 59, p. 142-157, 2015.
The 'APL.zip' file contains all APLs arranged by the 20 standard amino
acid residues (3 letter code) and eight different secondary structure
assigned by STRIDE.
Amino Acid 3-Letter-Code
Alanine Ala
Arginine Arg
Asparagine Asn
Aspartic acid Asp
Cysteine Cys
Glutamic acid Glu
Glutamine Gln Secondary Structure 1-Letter-Code
Glycine Gly Alpha helix H
Histidine His 3-10 helix G
Isoleucine Ile PI-helix I
Leucine Leu Extended conformation E
Lysine Lys Isolated bridge B or b
Methionine Met Turn T
Phenylalanine Phe Coil (none of the above) C
Proline Pro
Serine Ser
Threonine Thr
Tryptophan Trp
Tyrosine Tyr
Valine Val
Example:
File ALA_H_histogram.dat represents the APL for the Alanine (ALA) amino acid
with alpha-Helix (H) secondary structure.
In each *_histogram.dat file, we have 4 major groups:
PHI PSI Frequency {OMEGA}
When the amino acid residue has CHI angles they are represented as:
PHI PSI Frequency {OMEGA} {CHI}'s
Example:
ALA_H_histogram.dat
PHI PSI Freq. {OMEGA}
-94.000000 11.000000 0.000010 {0: [169.7]}
ASN_G_histogram.dat
PHI PSI Freq. {OMEGA} {CHI-1} {CHI-2}
-99.000000 11.000000 0.000240 {0: [-176.0]} {0: [-171.3]} {0: [48.1]}
When the number of occurrences grows, the number of {OMEGA} and {CHI}'s grows as well.
Example:
ALA_H_histogram.dat
PHI PSI Freq. {OMEGA}
-95.000000 20.000000 0.000019 {0: [172.6, 173.5]}
ASN_G_histogram.dat
PHI PSI Freq. {OMEGA} {CHI-1} {CHI-2}
-95.000000 -16.000000 0.000480 {0: [-175.5, -159.3]} {0: [-68.3, -66.1]} {0: [-51.5, -33.8]}
When the number of occurrences grows but the values of {OMEGA} or {CHI}'s
are different, sub-groups are made.
Example:
ALA_H_histogram.dat
PHI PSI Freq. {OMEGA}
-95.000000 -5.000000 0.000019 {0: [-179.1], 1: [172.0]}
ASN_G_histogram.dat
PHI PSI Freq. {OMEGA} {CHI-1} {CHI-2}
-95.000000 -11.000000 0.000480 {0: [-176.6, -163.6]} {0: [-79.8, -74.1]} {0: [-23.1], 1: [131.0]}
PHI PSI Freq. {OMEGA} {CHI-1} {CHI-2}
-91.000000 8.000000 0.000480 {0: [-179.2], 1: [173.4]} {0: [-76.7, -75.1]} {0: [-13.8, -9.4]}
PHI PSI Freq. {OMEGA} {CHI-1} {CHI-2}
-71.000000 -24.000000 0.000480 {0: [-176.1], 1: [175.3]} {0: [-170.2], 1: [-68.8]} {0: [-36.1], 1: [51.2]}